Optimization Tutorial¶
This tutorial describes mc3’s optimization function mc3.fit(),
which provides model-fitting optimization through scipy.optimize’s
leastsq (Levenberg-Marquardt) and least_squares (Trust Region
Reflective) routines. Additionally, the optimization can include
(two-sided) Gaussian priors, set shared parameters, and fixed
parameters. The mc3.fit() arguments work similarly to those of
the mc3.sample() function.
This function performs a Maximum-A-Posteriori optimization by
maximizing log_post (where we are neglecting the constant terms):
where the first term is the well known chi-square:
For the prior, the code computes log_prior as (once again
neglecting constant terms):
for each parameter with a Gaussian prior \(j\); parameters with
uniform priors do not contribute to log_prior.
Basic Fit¶
This is the function’s calling signature:
-
mc3.fit(data, uncert, func, params, indparams=[], indparams_dict={}, pstep=None, pmin=None, pmax=None, prior=None, priorlow=None, priorup=None, leastsq='lm')[source]
In the most basic form, the user only needs to provide the fitting
parameters and function, the data and \(1\sigma\)-uncertainties
arrays, (and any additional argument to the fitting function). This
will perform a Levenberg-Marquardt fit. The function returns a
dictionary containing the best-fitting parameters, chi-square, and
model, and the output from the scipy optimizer. See the example
below:
import numpy as np
import mc3
def quad(p, x):
"""Quadratic polynomial: y(x) = p0 + p1*x + p2*x^2"""
return p[0] + p[1]*x + p[2]*x**2.0
# Preamble (create a synthetic dataset, in a real scenario you would
# get your dataset from your own data analysis pipeline):
np.random.seed(10)
x = np.linspace(0, 10, 100)
p0 = [4.5, -2.4, 0.5]
y = quad(p0, x)
uncert = np.sqrt(np.abs(y))
data = y + np.random.normal(0, uncert)
# Fit the data:
output = mc3.fit(data, uncert, quad, [3.0, -2.0, 0.1], indparams=[x])
print(list(output.keys()))
['bestp', 'best_log_post', 'best_chisq', 'best_model', 'optimizer_res']
print(output['bestp'])
[ 4.57471072 -2.28357843 0.48341911]
# -2*log_post and chi-square are the same (uniform priors):
print(-2*output['best_log_post'], output['best_chisq'], sep='\n')
92.79923183159411
92.79923183159411
# Plot data and best-fitting model:
mc3.plots.modelfit(data, uncert, x, output['best_model'], nbins=100)
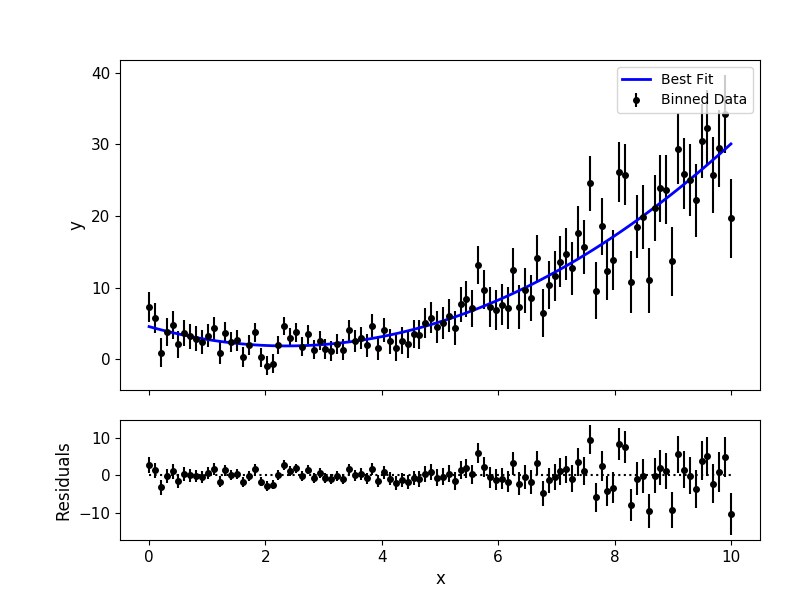
Data and Uncertainties¶
The data and uncert arguments are 1D arrays that set the data
and \(1\sigma\) uncertainties to be fit.
Modeling Function¶
The func argument is a callable that defines the parameterized
modeling function fitting the data. The only requirement for the
modeling function is that its arguments follow the same structure of
the callable in scipy.optimize.leastsq, i.e., the modeling
function has to able to be called as: model = func(params,
*indparams, **indparams_dict)
The params argument is a 1D array containing the initial-guess
values for the model fitting parameters.
The indparams and indparams_dict arguments (optional) set any
func additional argument as a list or keyword dictionary,
respectively.
Optimization Algorithm¶
Set leastsq='lm' to
use the Levenberg-Marquardt algorithm (default) via Scipy’s leastsq,
or set leastsq='trf' to use the Trust Region Reflective algorithm
via Scipy’s least_squares.
Fixed and shared-values apply during the optimization (see
Stepping Behavior), as well as the priors (see Parameter Priors).
Note
From the scipy documentation: Levenberg-Marquardt ‘doesn’t handle bounds’ but is ‘the most efficient method for small unconstrained problems’; whereas the Trust Region Reflective algorithm is a ‘Generally robust method, suitable for large sparse problems with bounds’.
The pmin and pmax arguments set the parameter lower and upper
boundaries for a trf optimization, e.g:
# Fit with the 'trf' algorithm and bounded parameter space:
output = mc3.fit(data, uncert, quad, [4.5, -2.5, 0.5], indparams=[x],
pmin=[4.4, -3.0, 0.4], pmax=[5.0, -2.0, 0.6], leastsq='trf')
Fixing and Sharing Paramerers¶
The pstep argument (optional) allows the user to keep fitting
parameters fixed or share their value with another parameter.
A positive pstep value leaves the parameter free, whereas a pstep
value of zero keeps the parameter fixed. For example:
# (Following on the script above)
# Fit the data, keeping the first parameter fixed at 4.5:
output = mc3.fit(data, uncert, quad, [4.5, -2.0, 0.1], indparams=[x],
pstep=[0.0, 1.0, 1.0])
print(output['bestp'])
[ 4.5 -2.24688721 0.47985918]
A parameter can share the value from another parameter by setting a
negative pstep, where the value of pstep is equal to the
negative index of the parameter to copy from. For example:
# (Though, it doesn't truly make sense for this model, let's pretend that the
# first and second parameters must have the same value, make a dataset for it:)
p1 = [4.5, 4.5, 0.5]
y1 = quad(p1, x)
uncert1 = np.sqrt(np.abs(y1))
data1 = y1 + np.random.normal(0, uncert1)
# Fit the data, enforcing the second parameter equal to the first one:
output = mc3.fit(data1, uncert1, quad, [3.0, -2.0, 0.1], indparams=[x],
pstep=[1.0, -1.0, 1.0])
print(output['bestp'])
[4.62479069 4.62479069 0.49179051]
Note
Consider that in this case, contrary to Python standards,
the pstep indexing starts counting from one instead of
zero (since negative zero is equal to zero).
Parameter Priors¶
The prior, priorlow, and priorup arguments (optional) set the
prior probability distributions of the fitting parameters.
Each of these arguments is a 1D float ndarray.
A priorlow value of zero (default) sets a uniform prior. This is
appropriate when there is no prior knowledge of the value of a
parameter \(\theta\):
Positive values of priorlow and priorup set a Gaussian prior.
This is typically used when a parameters has a previous estimate in
the form of \(p(\theta) = {\theta_p\,}^{+\sigma_{\rm
up}}_{-\sigma_\rm{lo}}\), where the
values of prior, priorlow and priorup define the prior
value, lower, and upper \(1\sigma\)
uncertainties, respectively:
where \(\sigma_{p}\) adopts the value of \(\sigma_{\rm lo}\) if \(\theta < \theta_p\), or \(\sigma_{\rm up}\) otherwise. The leading factor is given by \(A = 2/(\sqrt{2\pi}(\sigma_{\rm up}+\sigma_{\rm lo}))\) (see [Wallis2014]).
# (Following on the script above)
# Fit, imposing a Gaussian prior on the first parameter at 4.5 +/- 0.1,
# and leaving uniform priors for the rest:
prior = np.array([ 4.0, 0.0, 0.0])
priorlow = np.array([ 0.1, 0.0, 0.0])
priorup = np.array([ 0.1, 0.0, 0.0])
output = mc3.fit(data, uncert, quad, [3.0, -2.0, 0.1], indparams=[x],
prior=prior, priorlow=priorlow, priorup=priorup)
# Best-fit solution is dominated by the prior on the first parameter:
print(output['bestp'])
[ 4.01743461 -2.00989432 0.45686521]
# -2*log_post and chi-square now differ:
print(-2*output['best_log_post'], output['best_chisq'], sep='\n')
93.8012177730325
93.7708211946111